Innovative Structural Biology

Publications
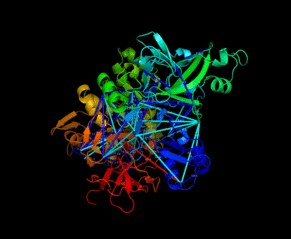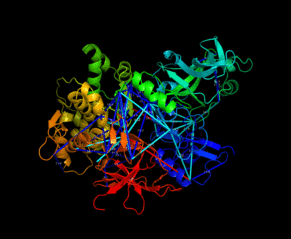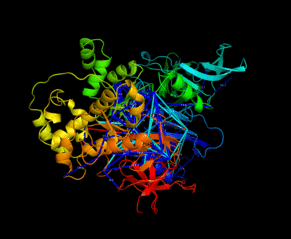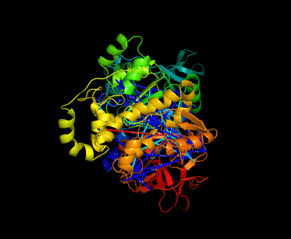
| Slavin M, Zamel J, Zohar K, Eliyahu T, Braitbard M, Brielle E, Baraz L, Stolovich-Rain M, Friedman A, Wolf DG, Rouvinski A, Linial M, Schneidman-Duhovny D, Kalisman N. Targeted in situ cross-linking mass spectrometry and integrative modeling reveal the architectures of three proteins from SARS-CoV-2. PNAS, 118:e2103554118, 2021. |
|
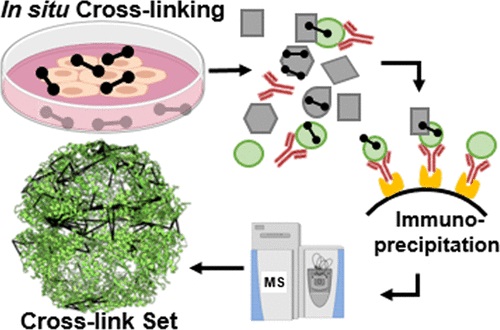
| Zamel J, Cohen S, Zohar K, Kalisman N. Facilitating In Situ Cross-Linking and Mass Spectrometry by Antibody-Based Protein Enrichment. J Proteome Res, 20:3701-3708, 2021. |
|
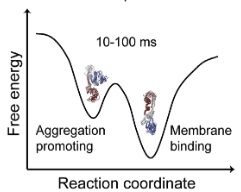
| Chen J, Zaer S, Drori P, Zamel J, Joron K, Kalisman N, Lerner E, Dokholyan NV. The structural heterogeneity of α-synuclein is governed by several distinct subpopulations with interconversion times slower than milliseconds. Structure, 29:1–17, 2021. |
|
| Leitner, et al. Toward Increased Reliability, Transparency, and Accessibility in Cross-linking Mass Spectrometry. Structure, 28:1259-1268, 2020. |
|
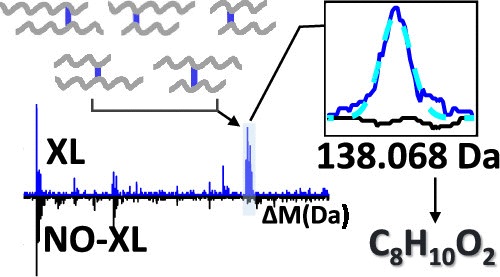
| Slavin M, Tayri-Wilk T, Milhem H, Kalisman N. Open Search Strategy for Inferring the Masses of Cross-Link Adducts on Proteins. Anal Chem, 92:15899-15907, 2020. |
|
| Tayri-Wilk T, Slavin M, Zamel J, Blass A, Cohen S, Motzik A, Sun X, Shalev DE, Ram O, Kalisman N. Mass spectrometry reveals the chemistry of formaldehyde cross-linking in structured proteins. Nat Commun, 11:3128, 2020. |
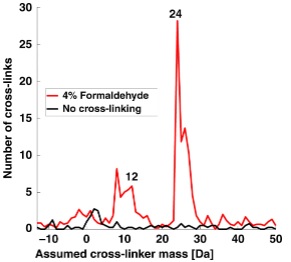
|
| Allu PK, Dawicki-McKenna JM, Van Eeuwen T, Slavin M, Braitbard M, Xu C, Kalisman N, Murakami K, Black BE. Structure of the Human Core Centromeric Nucleosome Complex. Curr Biol, 29:2625-2639, 2019. |
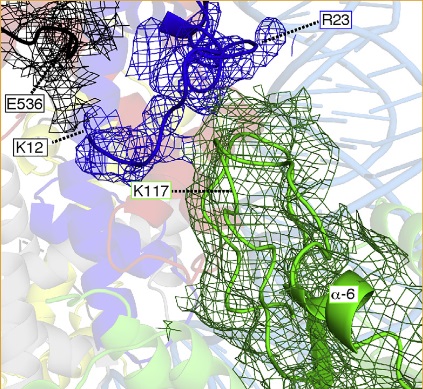
|
| Iacobucci C, et al. First Community-Wide, Comparative Cross-Linking Mass Spectrometry Study. Anal Chem, 91:6953-6961, 2019. |
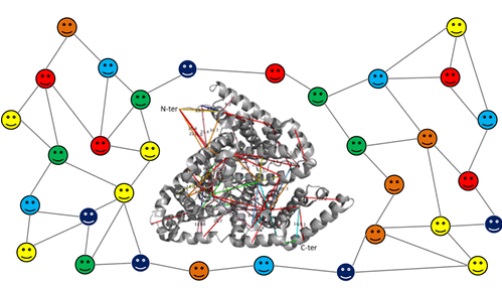
|
| Braitbard M, Schneidman-Duhovny D, Kalisman N. Integrative Structure Modeling: Overview and Assessment. Annu Rev Biochem, 88:113-135, 2019. |
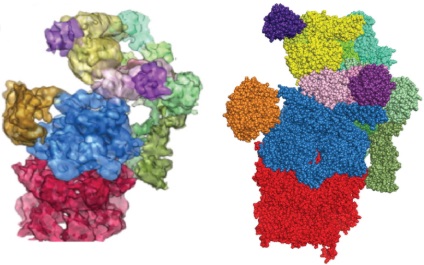
|
| Ben-Aharon Z, Levitt M, Kalisman N. Automatic Inference of Sequence from Low-Resolution Crystallographic Data. Structure, 26:1546–54, 2018. [PDF] |
|
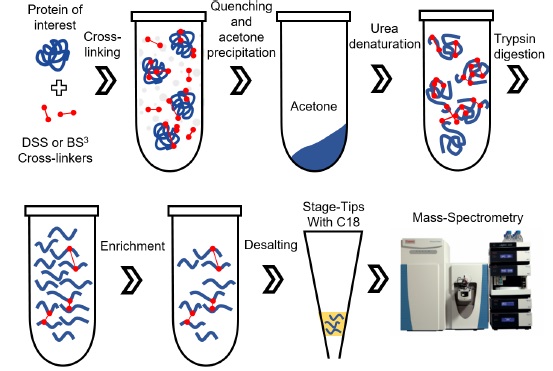
| Slavin M, Kalisman N. Structural Analysis of Protein Complexes by Cross-Linking and Mass Spectrometry. Methods Mol Biol, 1764:173-83, 2018. [PDF] |
|
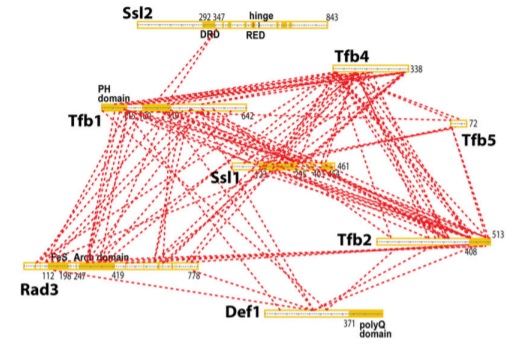
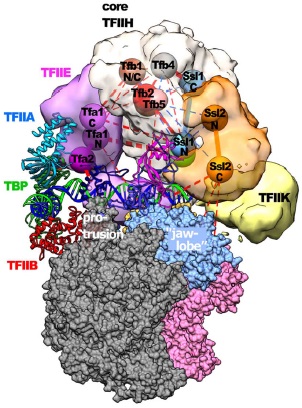
| Damodaren N, Van Eeuwen T, Zamel J, Lin-Shiao E, Kalisman N, Murakami K. Def1 interacts with TFIIH and modulates RNA polymerase II transcription. PNAS, 114:13230-5, 2017. [PDF] |
|
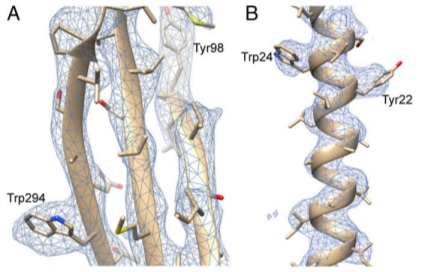
| Braun T, Vos MR, Kalisman N, Sherman NE, Rachel R, Wirth R, Schröder GF, Egelman EH. Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. PNAS, 113:10352-7, 2016. [PDF] |
|
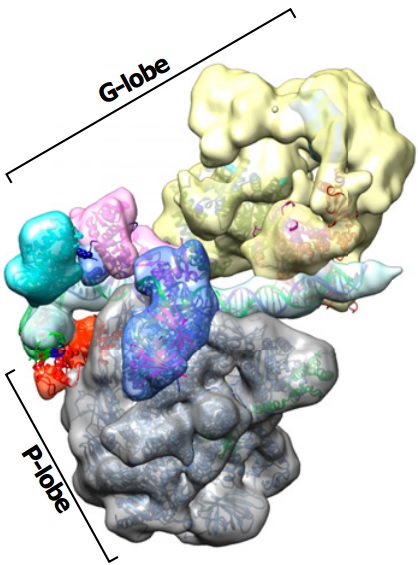
| Murakami K, Tsai KL, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD. Structure of an RNA polymerase II preinitiation complex. PNAS, 112:13543-8, 2015. [PDF] |
|
| Murakami K*, Elmlund H*, Kalisman N*, Bushnell DA, Adams CM, Azubel M, Elmlund D, Levi-Kalisman Y, Liu X, Levitt M, Kornberg RD. Architecture of an RNA polymerase II transcription pre-initiation complex. Science, 342(6159):1238724, 2013. [PDF] |
|
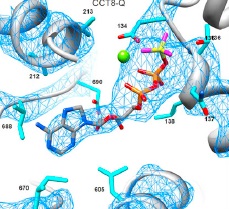
| Kalisman N, Schröder GF, Levitt M. The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning. Structure, 21:540-9, 2013. [PDF] |
|
| Kalisman N, Adams, CM, Levitt M. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. PNAS, 109:2884-9, 2012. [PDF] |

|
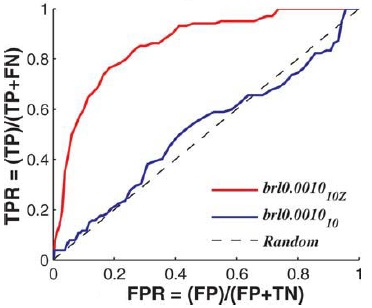
| Yahalom R, Reshef D, Wiener A, Frankel S, Kalisman N, Lerner B, Keasar C. Structure-based identification of catalytic residues. Proteins, 79:1952-63, 2011. [PDF] |
|
| Levi-Kalisman Y, Jadzinsky PD, Kalisman N, Tsunoyama H, Tsukuda T, Bushnell DA, Kornberg RD. Synthesis and characterization of Au102(p-MBA)44 nanoparticles. JACS, 133:2976-82, 2011. [PDF] |

|
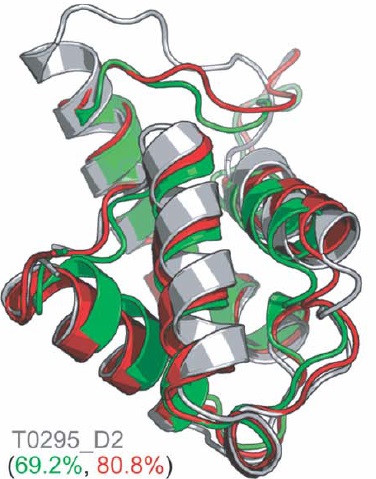
| Chopra G*, Kalisman N*, Levitt M. Consistent refinement of submitted models at CASP using a knowledge-based potential. Proteins, 78:2668-78, 2010. [PDF] |
|
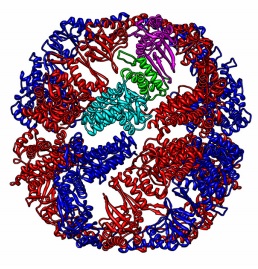
| Kalisman N, Levitt M. Insights into the intra-ring subunit order of tric/cct: a structural and evolutionary analysis. Pac Symp Biocomput, 252-9, 2010. [PDF] |
|
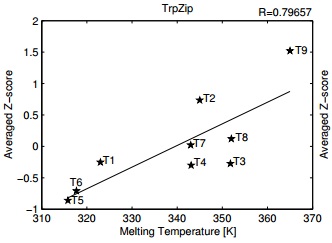
| Noy K, Kalisman N, Keasar C. Prediction of structural stability of short beta-hairpin peptides by molecular dynamics and knowledge-based potentials. BMC Struct Biol, 8:27, 2008. [PDF] |
|
| Amir ED*, Kalisman N*, Keasar C. Differentiable, multi-dimensional, knowledge-based energy terms for torsion angle probabilities and propensities. Proteins, 72:62-73, 2008. [PDF] |
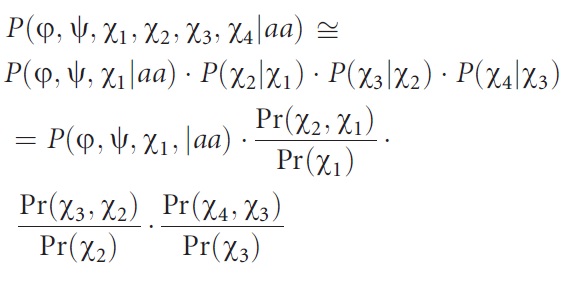
|
| Kalisman N, Levi A, Maximova T, Reshef D, Zafriri-Lynn S, Gleyzer Y, Keasar C. MESHI: a new library of Java classes for molecular modeling. Bioinformatics, 21:3931-2, 2005. [PDF] |
|
| Kalisman N*, Silberberg G*, Markram H. The neocortical microcircuit as a tabula rasa. PNAS, 102:880-5, 2005. [PDF] |
|
| Kalisman N, Silberberg G, Markram H. Deriving physical connectivity from neuronal morphology. Biol Cybern, 88:210-8, 2003. [PDF] |
|