Innovative Structural Biology

Research
How does cross-linking and mass-spectrometry (XL-MS) work?
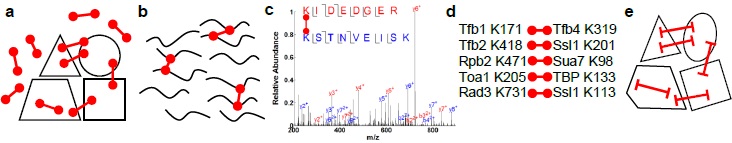
Cross-linking and mass-spectrometry is an emerging experimental approach that provides structural information
on protein complexes that are currently inaccessible by X-ray crystallography. The idea is to cross-link
the intact complex under native conditions with a short bi-functional cross-linking reagent (a). After the covalent
cross-linking occurred, the complex is denatured and digested with a protease (b). The digest is analyzed by mass-spectrometry (c),
and pairs of residues that underwent cross-linking are identified (d). Such analysis can identify hundreds of cross-linked
sites on a large complex. The cross-links are converted to distance constraints and drive computational modeling (e).
Studying the architectures of very large protein complexes with XL-MS
We recently used XL-MS to solve the architectures of two central eukaryotic complexes. The CCT chaperonin (16 subunits;
1 Mega-Dalton) is essential for the correct and efficient folding of certain cytosolic proteins. The transcription pre-initiation
complex (32 subunits; 1.5 Mega-Dalton) assembles before every round of transcription to correctly position the RNA polymerase
II enzyme on the DNA. The lab is employing XL-MS to expand these architectures by identifying new subunits that interact
with the complexes, and by obtaining more cross-links to refine the models. We have also started to probe the molecular
structure of the neuronal synapse with XL-MS.
Structural modeling with XL-MS data
Our lab is developing new computational ways to effectively convert XL-MS data into useful 3D models. The current focus
is on coarse-grained modeling, where each subunit is represented by a small set of points. We are working to merge this
level of modeling with other structural modalities such as SAX and cryo-Electron Microscopy.

Getting more out of low-resolution X-ray crystallography data
In collaboration with the lab of Michael Levitt at Stanford University, we are trying to find new computational
ways to use low-resolution crystallographic data more effectively. We have recently showed that crystallographic
data sets with resolutions as low as 5.5Å can single out the correct structural model out of millions of
alternative models. This finding, which directly relies on the mathematical fit between the model and the data,
opens the way to new uses of low-resolution crystallography.
